URGENT UPDATE: A groundbreaking study published on July 5, 2025, by a team at KU Leuven, reveals a revolutionary deep learning model that dramatically enhances the 3D imaging of fruit tissues, particularly apples and pears. This model outperforms traditional 2D methods, offering an unprecedented level of accuracy in understanding vital plant tissue microstructures, which are crucial for metabolic processes.
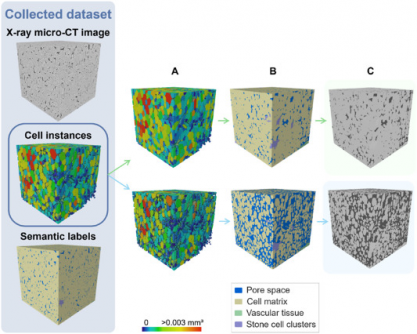
This research addresses long-standing challenges in plant tissue imaging, where traditional microscopy methods require extensive preparation and provide limited views. The innovative X-ray micro-CT technology allows non-destructive 3D imaging, yet previous methods struggled with complex tissue morphology. The new deep learning approach provides automated and precise segmentation of plant tissues, marking a significant advancement in the field.
The study, available in Plant Phenomics, introduces the first fully automated framework for labeling plant tissue architecture. The 3D panoptic segmentation framework utilizes advanced algorithms to accurately identify and classify various tissue types, including parenchyma cells and vascular structures. This model has achieved a remarkable Aggregated Jaccard Index (AJI) of 0.889 for apples and 0.773 for pears, surpassing previous benchmarks.
Key metrics from the study reveal that the model effectively segments pore spaces and cell matrices, achieving a Dice Similarity Coefficient (DSC) of up to 0.90 for certain tissue types. Specifically, it identified vascular tissues with DSC values of 0.506 for apples and 0.789 for pears, demonstrating a high level of precision in tissue analysis.
“This 3D deep learning model provides plant scientists with a powerful, non-destructive tool for studying how microscopic structures influence water, gas, and nutrient transport,” said Pieter Verboven, lead researcher from KU Leuven.
The implications of this research are profound. This model not only accelerates research into fruit storage and susceptibility to disorders like browning and watercore but also offers a scalable framework for studying plant development under various conditions. Its compatibility with standard X-ray micro-CT instruments makes it an accessible solution for integrating artificial intelligence into plant anatomy and food science research.
As the agricultural sector increasingly seeks innovative solutions to enhance crop resilience and quality, this technology could transform how researchers approach plant physiology. The deep learning model drastically reduces manual labor in tissue characterization while improving accuracy, paving the way for faster, more efficient studies in the field.
Moving forward, the research team will explore further enhancements and potential applications of this technology in various crops. The results from this study highlight not only the advancements in imaging techniques but also the urgent need for innovative tools to address global agricultural challenges.
Stay tuned for more updates as this technology continues to develop and reshape the future of plant research.