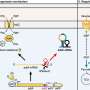
A recent study has revealed how the bacterium Salmonella employs a small RNA derived from its 3′ untranslated region (3′UTR) to connect its capacity for acid resistance with metabolic reprogramming while residing within macrophages. This discovery sheds light on the adaptive strategies of pathogens and their survival mechanisms in hostile environments, particularly during infection.
Acid resistance is essential for enterobacteria like Salmonella as it enables them to endure the acidic conditions found in the host’s gastrointestinal tract and within macrophage phagosomes. These conditions are typically hostile to many microorganisms, making the ability to resist acidity a critical factor for pathogen survival.
One of the key components in this process is the enzyme arginine decarboxylase AdiA. This enzyme plays a significant role in acid resistance by facilitating a reaction that consumes protons (H+), thereby helping to neutralize acidic environments. The research provides insight into how this enzymatic activity is regulated by the small RNA, which appears to be integral to Salmonella’s metabolic adjustments.
Research Findings and Implications
The study highlights the complex interplay between acid resistance and metabolic pathways in Salmonella. The small RNA not only aids in adapting to acidic conditions but also influences overall metabolic reprogramming. This dual role demonstrates how pathogens can efficiently manage their survival strategies in the face of host defenses.
Furthermore, understanding the mechanisms behind acid resistance could open pathways for the development of new therapeutic strategies aimed at combating infections caused by Salmonella and similar pathogens. By targeting the metabolic pathways linked to acid resistance, researchers may find novel approaches to inhibit bacterial growth and improve treatment outcomes.
The significance of these findings extends beyond just understanding Salmonella. They underscore the importance of metabolic flexibility in pathogens and how this flexibility contributes to their virulence. As researchers continue to explore these mechanisms, it may become possible to develop more effective interventions against a range of bacterial infections.
In conclusion, the discovery of the role played by 3′UTR-derived small RNA in linking acid resistance to metabolic reprogramming in Salmonella within macrophages not only enhances our understanding of microbial survival strategies but also highlights the potential for new therapeutic targets in the fight against bacterial infections.